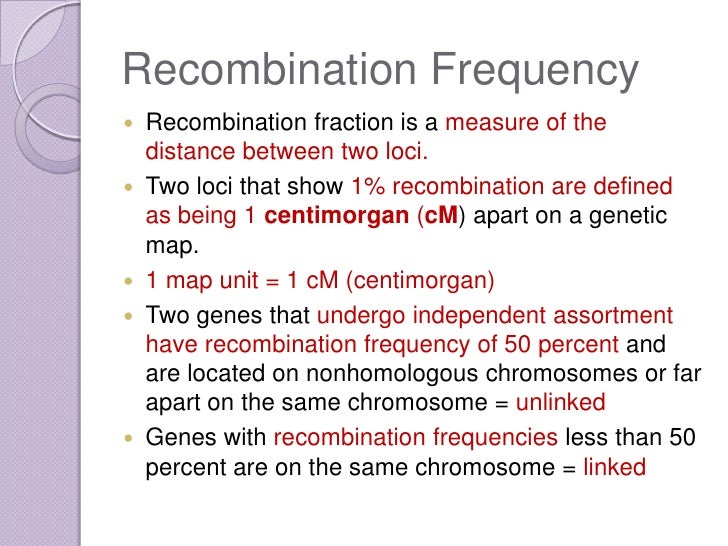
Calculating recombination frequency map units
Once we've assigned our mutation to a linkage group, it's generally off to the races with three-point and SNP mapping methods, as these will almost always be necessary to clone our genes anyway. It is worth noting, however, that high-throughput methods for two-point mapping using SNPs also see below have been used successfully by some groups and can provide a very precise map position for mutations.
These methods may even in some cases allow for the molecular cloning of mutations in the absence of further three-point or SNP mapping Wicks et al. Nevertheless, the vast majority of researchers still use the standard tiered cloning progression for which two-point mapping is simply step one.
In carrying out two-point mapping, one can use either genetic or SNP markers. For the purpose of this section, we will cover the use of genetic markers, as the principals are the same and may be somewhat easier to grasp.
Often we will carry out traditional two-point mapping using strains that contain two adjacent chromosomal markers, as the strains generated by these crosses can potentially be used later for three-point genetic mapping. The two most basic outcomes for two-point mapping are shown in Figure 1. In this case, the mutation is actually flanked by the markers to produce a reasonably well-balanced strain.
The genotypes of the progeny are indicated along with the ratios or fractions of their occurrences. In this situation we essentially never see the appearance of the triple mutant phenotype MAB , as this would require an exceedingly rare double-recombination event to take place.
Furthermore, if we were to pick animals of phenotype M and examine their self-progeny, we would never see M A B animals. Finally, wild-type animals will always throw both M and A B along with wild-type animals. Seeing segregation patterns of this type tells us that m and a b reside on the same chromosome and also that m resides close to or in between the markers a and b.
In the circumstance that the M phenotype is lethal, this may be a useful strain for maintaining m as a balanced heterozygote. Namely, by isolating wild-type segregants at each generation, we can propagate the mutation with relative ease.
In addition, this strain can be used for three-point mapping. In contrast, the situation depicted in scheme 2 shows m and a b on distinct chromosomes. If necessary, draw out all the possible genotypes and corresponding phenotypes to convince yourself that these numbers are correct. Observing these kinds of segregation patterns indicates that the mutation and the markers are on different chromosomes. Another possibility is that the mutation resides on one of the ends of the chromosome see below.
If necessary, these two possibilities can usually be resolved by scoring more animals. In general, basing linkage designation on a small number of data points fewer than 20 should be avoided. The genetic patterns described above are for the ideal situation where there is no ambiguity in the determination of chromosomal location.
But what happens when the mutation lies to one side of the markers, perhaps at some distance? As shown in Figure 2 , if the mutation lies to one side, a cross over may occur that will lead to the creation of the two recombinant chromosomes shown. One recombinant chromosome will now contain all three mutations in cis, whereas the other is completely wild type. Also shown are the genotypes occurring when such a recombinant chromosome is paired with one of the parental chromosomes.
In addition, a wild-type animal can now fail to throw both M and AB animals. In thinking about this, keep in mind that these "rare" recombinant chromosomes will usually - by chance - wind up paired with one of the non-recombinant parental chromosomes in a fertilized zygote. Of course, the farther the mutation is from the markers, the higher the proportion of recombinant chromosomes in the pool, and the greater the possibility that any two recombinant chromosomes may end up together in a zygote.
In these situations we must be careful not to hastily conclude that the presence of such genotypes automatically means that m and a b are on separate chromosomes. Genetic Analysis of Single Genes Chapter 4: Mutation and Variation Chapter 5: Pedigrees and Populations Chapter 6: Genetic Analysis of Multiple Genes Chapter 7: Linkage and Mapping Chapter 8: Techniques of Molecular Biology Chapter 9: Changes in Chromosome Number and Structure Chapter Molecular Markers and Quantitative Traits Chapter Genomics and Systems Biology Chapter Regulation of Gene Expression Chapter Main Discussion Cube 0.
Login to add a new comment. The method for mapping of these long chromosomes is shown below. Note that the map distance of two loci alone does not tell us anything about the orientation of these loci relative to other features, such as centromeres or telomeres, on the chromosome.
Map distances are always calculated for one pair of loci at a time. However, by combining the results of multiple calculations, a genetic map of many loci on a chromosome can be produced Figure 7. A genetic map shows the map distance, in cM, that separates any two loci, and the position of these loci relative to all other mapped loci. The genetic map distance is roughly proportional to the physical distance, i.
For example, in Arabidopsis , 1. When a novel gene or locus is identified by mutation or polymorphism, its approximate position on a chromosome can be determined by crossing it with previously mapped genes, and then calculating the recombination frequency. If the novel gene and the previously mapped genes show complete or partial linkage, the recombination frequency will indicate the approximate position of the novel gene within the genetic map.
This information is useful in isolating i. Genetic maps are useful for showing the order of loci along a chromosome, but the distances are only an approximation.
The correlation between recombination frequency and actual chromosomal distance is more accurate for short distances low RF values than long distances.